Table of Contents
ToggleIsolation of Genomic DNA from bacteria is a little tedious and time-consuming methods but a useful and crucial method for the isolation of bacteria from soil and helps in characterizing the soil bacteria. As we know that DNA is the main part of life and can be so useful for the identification of organisms’ nucleotides.
First, the important part is to isolate pure culture from the bacterial colonies by a different method that we discussed in our previous article, then only we can isolate Genomic DNA from the bacteria by different methods. We are going to discuss the Phenol chloroform method for this process. A variety of chemicals and enzymes were used in this specific method. We will be going to explain all the functions of those chemicals too.
Isolation of Genomic DNA from bacteria principle
The fundamental method of isolating DNA involves breaking down the cell wall, nuclear membrane, and cell membrane to release the highly intact DNA into solution. Next, DNA is precipitated while contaminating biomolecules like proteins, polysaccharides, lipids, phenols, and other secondary metabolites are removed.
Isolation of Genomic DNA from bacteria protocol:
Genomic DNA was isolated from bacteria by Phenol and chloroform method.
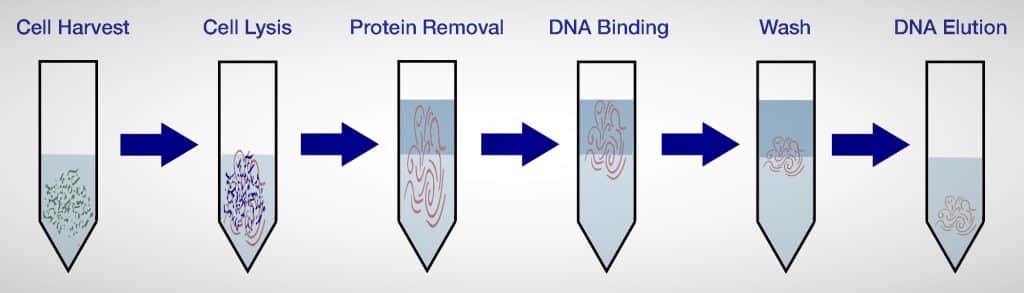
- First of all, we have to isolate a pure culture of bacteria by different methods, and then only after seed culture, a small part will be taken for fermentation for extracting secondary metabolites.
- And another part of the seed culture about 15mL of pure cultures was taken in a falcon tube.
- After centrifugation at about 5 minutes at 8000 rpm and the supernatant was removed.
- Cell pellets were collected and the process of isolation of genomic DNA was started.
- Harvested cell pellets were then lysis with lysis buffer that is 10% sucrose and was vertex strongly followed by centrifugation and this process was repeated 2 times.
- The supernatant was removed and the pellets were mixed with 240 microliters of 8mg/mL lysozyme and the sample was kept at 37 degrees Celcius.
- After complete lysis, 240 microlitres of 0.5M EDTA and 26 microlitres of 10 mg/mL proteinase K were added.
- The samples were again kept in a water bath for 5 minutes at 37 degrees Celcius.
- SDS (10%) 200 microlitres was added and samples were kept at 70 degrees Celcius for 10 minutes and were immediately transferred to an ice bath for about 15 minutes.
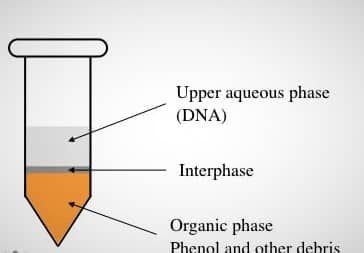
- Then an equal volume of Phenol: Chloroform: isoamyl alcohol reagent (25:24:1) was added to the samples and was shaken gently, and centrifuged at 8000 rpm for 5 minutes.
- Now the three layers were observed, the upper layer was aqueous in which cell DNA was present, the middle interphase, and the lower one is an organic layer. The supernatant was transferred into a new Eppendorf tube without disturbing the solvent layer.
- Then, 40 microlitres of 1mg/mL RNase was added and kept at 37 degrees Celcius for an hour.
- After all these enzymes and chemicals, 0.8 volumes of isopropanol and 2 volumes of 100 % alcohol were added into the tube and shaken slowly.
- For the precipitation of DNA, the tubes were incubated at -20 degrees Celcius for 30 minutes.
- After this process, the solution was centrifuged and washed with 70% ethanol and the remaining ethanol was evaporated by incubation at room temperature overnight.
- Finally, after putting it overnight, 10 microlitres of tris-base-EDTA (TE) buffer was added to the tubes and we get the DNA after centrifugation at 5000rpm for 10 seconds.
- Now, the genomic DNA was examined through Gel electrophoresis.
Chemicals used during isolation and their function:
The various chemicals used during the isolation of genomic DNA are:
- Tris-buffer- It generally maintains the Ph of the solution and helps in making the membrane permeable resulting in lysis.
- EDTA- A chelating agent, chelates the magnesium ions which act as a cofactor for the enzyme.
- SDS- Act as detergents, which helps in the lysis by removing lipid molecules and thereby causing disruption of cell membranes.
- Acetate- Protect from denaturation
- Phenol- The addition of phenol breaks the bond between amino acids leading to the denaturation of protein.
- Chloroform- Allow proper separation of the organic and aqueous phases.
- Isoamyl alcohol- It prevents the emulsification of the solution.
MCQs/FAQs
Which solvent is used in DNA extraction?
Phenol: chloroform: isoamyl alcohol is used as a standard solvent in DNA extraction.
Why is Phenol chloroform used in DNA extraction?
The aqueous phase containing nucleic acids can subsequently be extracted. Chloroform is frequently used in conjunction with phenol. To establish a distinct separation between the aqueous and organic phases, chloroform is added along with phenol. Contrary to phenol and water, chloroform and phenol are miscible.
What are the three basic steps in the extraction of DNA?
Step one of the lysis process involves rupturing the cell and nucleus to liberate the DNA.
Precipitation (Step 2): Following lysis, DNA has been released from the nucleus but is still intermingled with fragments of the cell.
Step three of the purification process resulted in the separation of the DNA from the aqueous phase.
What is the purpose of DNA extraction?
For the development of diagnostics and medications as well as for research into the genetic causes of disease, the capacity to extract DNA is of utmost significance. Performing forensic science, sequencing genomes, spotting bacteria and viruses in the environment, and establishing paternity are all made possible by it.
Why is alcohol is used in DNA extraction?
For the development of diagnostics and medications as well as for research into the genetic causes of disease, the capacity to extract DNA is of utmost significance. Performing forensic science, sequencing genomes, spotting bacteria and viruses in the environment, and establishing paternity are all made possible by it.
What is the use of SDS in DNA extraction?
In laboratories, SDS is frequently used as a component of the buffer for cell lysis, cell lysis during DNA extraction, and primarily in the running buffer for SDS-PAGE. SDS is an anionic detergent that is given to a protein sample to linearize the proteins and give them a negative charge.
Why is proteinase k is used in DNA extraction?
Since proteinase K effectively inactivates DNases and RNases, it is used to break down proteins in cell lysates (tissue, cell culture cells) and to release nucleic acids.
Why is EDTA used during DNA extraction?
DNAse, an enzyme that cleaves DNA by dissolving its phosphodiester links, is activated by these metal ions. The main reason EDTA is employed in the purification steps of DNA analysis is to remove these metal ions to prevent DNAse from functioning.