Table of Contents
ToggleIn this blog, we have tried to explain the different techniques for the isolation of bacteria including various steps.
Isolation of bacteria from soil
Isolation of bacteria can be originated from the soil, while some are also isolated from water and the air. One spoon of soil contains 1,000 bacteria, and around 90% of all bacteria are isolated from it. Multiple methods are used for the isolation of bacteria from soil. Bacteria that are present in various harsh environments develop various forms of capacity to combat them as well as various components known as secondary metabolites, sometimes known as antibiotics, which bring new innovations to mankind. Producing antibiotics from soil bacteria is a highly intriguing and special technique that can be quite helpful for us. The majority of them are gram-positive actinomycetes that have been cultivated in a lab.
There is a variety of techniques to isolate bacteria from air, soil, and water. Let us jump into the isolation of bacteria ( Streptomyces species) from the soil.
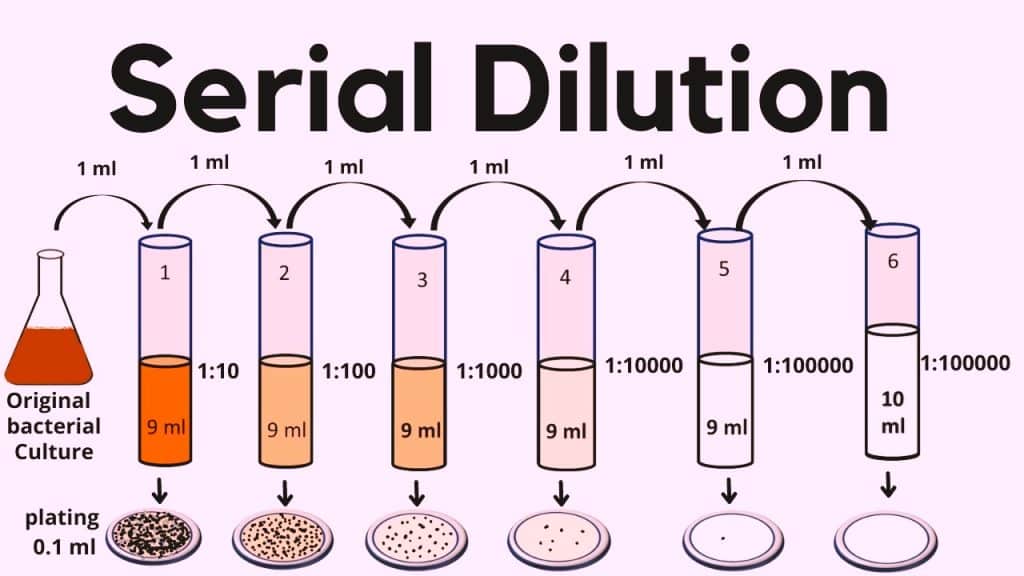
Isolation of bacteria by dilution method
Serial dilutions (also known as dilution method) are used in microbiology to lower bacterial concentrations to the levels necessary for a given test method or to levels that make it simpler to count bacteria when they are plated onto an agar plate. The major steps involved in this particular method are listed below:
- Collection of soil
- Isolation of actinomycetes
- pour plate methods
- Purification of pure culture
- Isolation of genomic DNA from bacteria
- Purity examination by gel electrophoresis
- Flask fermentation
- Ethyl acetate extraction
- Identification of bacteria by Gram’s staining techniques
- lC-HRMS of bacterial extracts
Collection of soil
Soil samples can be collected from different parts of the country from 2-3 inches approx. 10-12.5 cm below the earth’s surface, where the majority of antimicrobial properties originate and the bacterial community is dense. With these a sterile spatula, plastic bag, and rubber band, ~500g soil samples will be gathered from different sites varying in soil color, moisture, organic matter particle sizes, and latitude and will be stored at 4°C in the lab, until further use, to avoid the contamination.

Isolation of actinomycetes
1gm of each soil sample will be added to 9mL autoclaved water so to make the volume 10mL and subjected to 80°C for 30 minutes to destroy vegetative cells. On sterilized water, the samples will be serially diluted up to two folds (0.001) concentration. For isolation, 100 µL(0.1mL) of suspension will be spread over ISP4 media, and 20mg/mL Nalidixic acid and 50 mg/ml cycloheximide will be added to the media to inhibit gram-negative bacterial and fungal growth, respectively. The plates will be incubated at 28°C for 5-8 days.
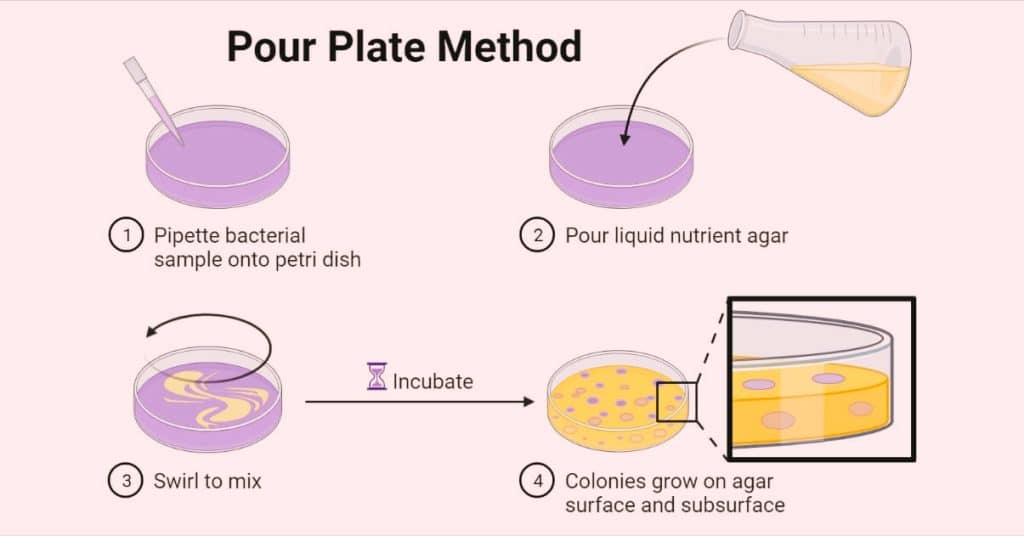
Pour plate methods
The pour plate technique is frequently used for bacteria, however, it is rarely utilized for fungus or actinomycetes. The original inoculum is diluted repeatedly, then placed in sterile Petri plates with melted, cooled (45°C), and thoroughly mixed (by rotating) agar media, and incubated after solidification.
Purification of pure culture
To obtain pure culture, previously grown bacterial colonies will be streaked on ISP4 media and incubated for 7-14 days at 28°C.

Isolation of genomic DNA from bacteria
The single colony will be cultured for 4-5 days at 180 rpm in TSB containing different flasks with glass beads at 28 ̊C. Cell platelets will be extracted from 15mL culture and taken in a falcon tube by centrifugation at 8000 rpm for 16 minutes. Thus obtained cell platelets will be treated with 3mL cell lysis buffer and 10% sucrose, centrifuged twice to remove the supernatant, and samples will be maintained at 37°C for 1 hour after adding a small amount of lysozyme.
Following the completion of lysis, 240 µL of 0.5M EDTA and 26 µL proteinase k (10mg/mL) will be added and kept at 37°C for 5 minutes. After adding 200 µL of 10% SDS, the sample will be kept at 70 for 10 minutes, then 500 µL of 5M potassium acetate will be added to it and placed in an ice bath for 15 minutes. Phenol, chloroform, and isoamyl alcohol will then be mixed in the ratio of 25:24:1 and centrifuged at 4000rpm for 10 minutes. The supernatant will be moved into new tubes and 0.5 volume of chloroform will be added and centrifuged at 4000rpm for 10 minutes.
Now, To the supernatant is transferred to a new tube, and 40 µL RNase will be added and kept at 37 ̊C for an hour. 0.8 volume of isopropanol and 2 volume of 100% ethanol will be added to the tube and shaken gently and incubated at -20 ̊C for an hour when DNA precipitates.
Then, the tube will be centrifuged again, ethanol will be removed and the DNA pellets obtained will be washed twice with 70% ethanol and dried at room temperature. Finally, the dried DNA will be dissolved in a TE buffer.
Purity examination by gel electrophoresis
The local purity of DNA will be checked by 0.4% agarose gel electrophoresis with the addition of 10mg/mL ethidium bromide (EtBr) as a fluorescent agent. The isolated genomic DNA will be mixed with an equal volume of 6X loading dye in 1:1 proportion and will be loaded in an agar well. 1 kb DNA ladder will also be loaded in a separate well.
Electrophoresis will be carried out at 50 V for 90 minutes in a 1X TAE solution. In Gel Doc, the DNA band and intensity will be compared to a 1 kb DNA ladder. DNA amplification and purification16s rDNA will be amplified by PCR using the set of universal primer, fwd_ name: 27F, fwd_ seq: AGAGTTTGATCMTGGCTCAG, rev _name: 1492R, rev _seq: ACGGYTACCTTGTTACGACTT. The primer will be diluted to a working concentration of 10 µM and the amplification will be carried out in a 50 µL reaction mixture containing Taq-polymerase enzyme within 35 cycles and 51.4 annealing temperature.
16s rRNA amplification will be examined by performing gel electrophoresis. The PCR product will be purified using a purification kit, the purified product will be subjected to electrophoresis and the product will be sent to South Korea for its sequencing.
Flask fermentation
Individual isolates will be cultured in TSB media for 3-4 days at 28°C and 140rpm in a shaking incubator. Following proper bacterial growth, 1% of the bacterial solution will be transferred to TSB at 28°C for 5-7 days at 140 rpm.
Ethyl acetate extraction
The fermented solution will then be mixed with an equal volume of ethyl acetate and incubated overnight in a shaking incubator. Then, the mixture will be centrifuged to get a supernatant that will be used for screening of antibacterial activity as an extract.

Identification of bacteria by Gram’s staining techniques
Gram staining techniques are still used today to distinguish between Gram-positive and Gram-negative microorganisms based on their color. Gram stains function as differential stains because some bacteria retain a primary stain (crystal violet) to withstand decolorization.

Screening and evaluation of antibacterial activity
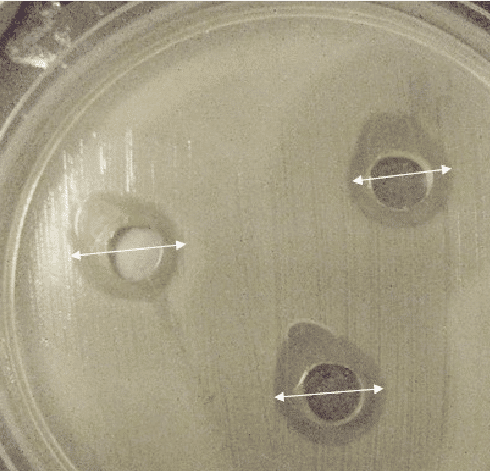
Agar-well diffusion technique will be used to test the extract’s antibacterial activity. The ethyl acetate extract will be carpet cultured with cotton swabs on MHA. Using a sterile cork borer, 6mm wells will be made and extract will be loaded using a micropipette. As a reference point, neomycin will be employed. Then it will be incubated at 37°C for 24 hours. After 24 hours, the zone of inhibition will be observed and measured using a ruler.
LC-HRMS of bacterial extracts
Based on the zone of inhibition values, the extracts were subjected to LC–HRMS which was carried out at Sophisticated Analytical Instrument Facility (SAIF), CSIR-Central Drug Research Institute, Lucknow. An Agilent 6520, Accurate-Mass Q-TOF Mass Spectrometer with a G1311A quaternary pump, G1329A autosampler, and G1315D diode array detector was used to examine the samples (DAD). The utilized particle size was 4. Acetonitrile (ACN), water, and a 5 mM acetate buffer were used in a gradient system for the solvent elution over the course of 40 min.
Starting with 5% ACN for 0.1 min, the initial condition increased to 30% ACN for 10 min, 80% ACN for 32 min, and then returned to its initial conditions. The column temperature was held constant throughout at 30 °C. The column elute was directed to Q-TOF HRMS equipped with an electrospray interface after passing through the DAD flow cell (ESI).
The raw data of LC-HRMS were analyzed through Mestre Nova 12.0 software for peak detection, alignment, and annotation and compared to the database library, literature, dictionary of natural products, and METLIN metabolite searching cloud.
FAQs/MCQs
What is the isolation of soil?
Techniques for isolating specific cells from a complex mixture of cells include cell identification, cell separation, and cell transfer. The goal is to get single cells or sort the cells based on the desired attribute and produce a population of cells that is homogeneous.
Which media is used during the isolation of bacteria?
ISP( international Streptomyces project) is used during the isolation of bacteria.
Which media is used during seed culture and fermentation?
TSB( tryptone soya broth) is used during fermentation.
Which media is used during the evaluation of antibacterial activity?
MHA( Muller Hinton agar) is used during the evaluation of the antibacterial activity.
What is the purpose of ethyl acetate during fermentation?
The purpose of ethyl acetate is to burst the cell of bacteria and release secondary metabolites from it.
Which technique is used during the screening of antibacterial activity?
Agar well diffusion is used to screen the antibacterial activity.