“How to perform molecular docking” is the commonly encountered query in drug design research. Molecular docking is a technique used to predict the binding energy and interaction of ligands with amino acid residues of protein. This computer-aided drug design technique is widely used in drug discovery. Docking is a helpful tool for lead optimization since it can be used to virtually screen huge compound libraries, and suggest structural ideas about how the ligands inhibit the target.
There are several molecular docking software. Autodock Vina is one of the most cited docking tools to date. Here are the protocols for performing molecular docking in drug discovery via Autodock Vina.
Tools for Autodock Vina
Steps in Molecular Docking
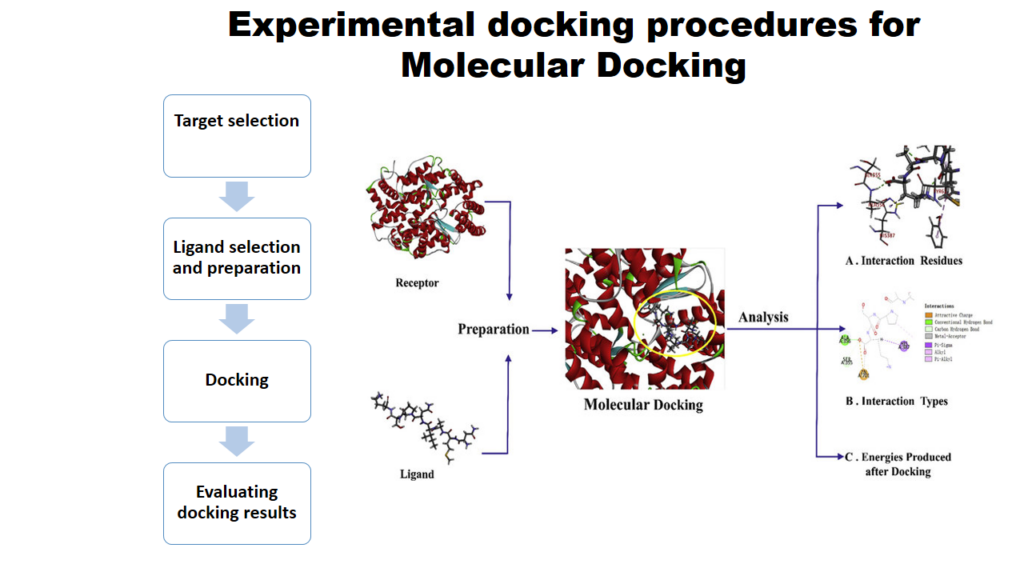
- Selection and preparation of ligand
- Go to PubChem and search ligand name
- Download ligand into 3D SDF format
- Open the ligand through Discovery Studio and save it into PDB format naming ligand
- Open Autodock vina and drag the PDB format ligand into it
- Click on the ligand option
- Go to input → choose select ligand
- select for Autodock
- Go to output and save into PDBQT format
- Selection and preparation of Protein
- Go to RCSB PDB and search protein
- Download protein of good resolution in PDB format
- Open the protein in Discovery Studio, delete unwanted chains & heteroatoms, and save it into PDB format naming the protein
- Drag the protein in Autodock Vina
- Go to Edit → Delete Water
- Go to Edit → Add Polar Hydrogen
- Go to Edit → Add Kollmann Charge
- Go to select → select from string → Insert amino acid residue (Active site)(Example: ASP197,GLU233)
- Go to Grid → macromolecules→ choose → protein → save into PDBQT
- Go to Grid → Grid box → Arrange grid box enclosing active site residues and save the grid txt file
- Prepare config.txt file as

- Run Command for molecular docking
- Copy file path
- Search “cmd” in the taskbar and perform the following command:
- cd (paste file location) and then press enter
- vina.exe –help (press enter)
- vina.exe –config config.txt –log log.txt (press enter)
- vina_split.exe –input output.pdbqt
- Analyze the binding energy results generated in log file
- Interaction results analysis by Discovery Studio
- Drag protein and 9 generated output ligands into discovery studio
- Select one protein.pdbqt and one ligand.pdbqt
- Go to define the receptor and ligand
- Go to define ligand → ligand interactions → Hydrogen bond
- Go to show 2D diagram
- View the interactions
- Select other ligands and perform the same process to view their interactions.
- Choose the one ligand output that showed the highest number of H-bonding with active site residue of the protein.
- Validation of docking protocol
- Select the best pose with the highest number of hydrogen bonding with the active site.
- Re-dock the ligand in the same pocket of the protein and choose the best-docked pose
- Drag best-docked pose of docking and redocked pose in Discovery studio
- Go to Structure →superimpose → Molecular overlay
- Go to Structure →RMSD → All atoms
- RSMD value of less than 2Å verifies the docking protocol