Table of Contents
ToggleSouthern Blotting is a laboratory procedure used to identify particular DNA sequences in a sample. It bears the name of its inventor, Dr. Edwin Southern, who created the technique in 1975. Southern blotting is frequently used in molecular biology research to measure the size of a DNA fragment, detect the presence or absence of a certain DNA sequence, and examine the chromosomal or gene structure. It can be used for gene expression analysis, DNA fingerprinting, genetic testing, and the research of hereditary illnesses.
Southern Blotting Definition
The Southern Blotting method involves the separation of DNA fragments using gel electrophoresis on an agarose gel. The DNA is subsequently capillary- or vacuum-transferred (or “blotted”) onto a membrane, often one made of nylon or nitrocellulose. A tagged DNA or RNA probe complementary to the target sequence of interest can be used to probe the immobilized DNA on the membrane. Autoradiography, fluorescence, or chemiluminescence can be used to detect the bound probe once the probe hybridizes with the membrane’s target sequence.
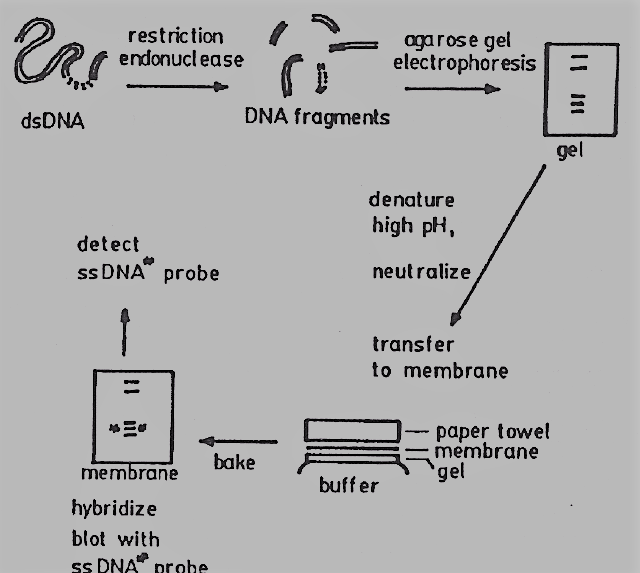
Southern Blotting Principle
The basis of Southern blotting is the separation of DNA fragments by gel electrophoresis, followed by their detection by tagged probe hybridization. During electrophoresis, the DNA fragments are divided based on their size and charge. The process entails moving electrophoresed DNA from an electrophoresis gel to a nitrocellulose membrane. The transfer buffer is drawn from the gel through capillary action by the nitrocellulose filter, which is placed between the gel and the stack of paper towels. Utilizing a labeled probe complementary to the desired DNA, the desired DNA is found.
Southern Blotting Protocol
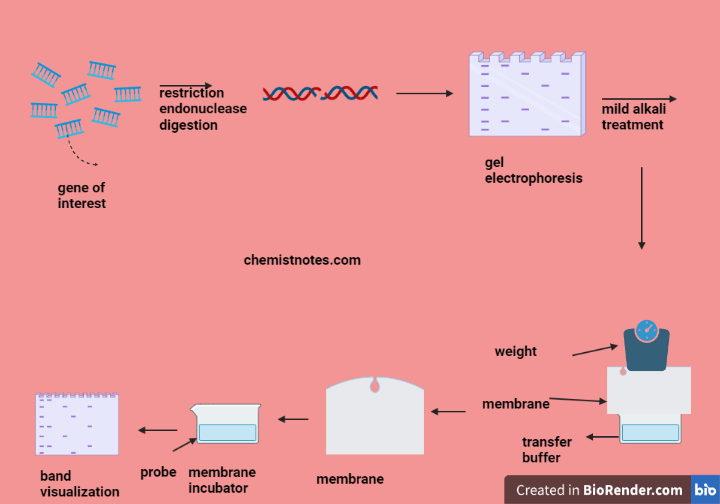
- DNA extraction: From the sample (such as tissue, blood, or cells), DNA is isolated. The technique produces high molecular weight DNA while preventing DNA deterioration is used.
- Restriction digestion enzyme: When restriction enzymes are used to cut DNA, they produce fragments of the desired size range for your research. When it comes to buffer and enzyme quantities and recommended incubation times, always abide by the manufacturer’s instructions.
- Gel electrophoresis: To separate the fragments by size, the digested DNA fragments are loaded onto an agarose gel and electrophoresis is run. To determine the sizes of the fragments, include a DNA ladder. To see the bands, the gel with ethidium bromide or another DNA dye is stained.
- Transfer of membrane: A piece of nylon or nitrocellulose membrane is cut to the gel’s size. The membrane on top of the gel is placed after soaking it in the transfer buffer. The membrane/gel stack is subjected to even pressure using a stack of paper towels or a sponge. The DNA to the membrane using electrophoresis is transferred. A current for one to two hours using a power supply, with the cathode (-) at the membrane and the anode (+) at the gel is applied.
- Hybridization: By Using a technique that labels the probe with a recognizable marker (such as radioactivity, fluorescence, or biotin), the label is probed. A hybridization buffer is used to dilute the probe before incubating the membrane with it at 42°C for an entire night. The corresponding DNA sequences on the membrane are bound to the probe.
- Washing: By using a wash buffer to clean the membrane, we may get rid of the unbound probe. A number of washes in succession with escalating rigor is used (for example, 2x SSC, 0.1% SDS, 65°C; 0.1x SSC, 0.1% SDS, 65°C).
- Detection: The membrane is exposed to detection chemicals designed to work with the probe’s particular label. This will produce a signal that can be seen by chemiluminescence, fluorescence, or autoradiography.
- Analysis: The membrane’s band pattern is analyzed to the expected pattern based on our hypothesis to analyze the results. By comparing the intensity of the bands to a standard curve formed with known amounts of DNA, and may calculate the amount of target DNA that is present in our sample.
Southern Blotting Application
If a gene is amplified, deleted, or physically altered in cancer cells as opposed to healthy cells, it can be determined using a Southern blot analysis. Although labor-intensive, this method is especially helpful for finding significant deletions in tumor genomes.
The DNA analysis method known as Southern blotting is widely used in molecular biology. It has been utilized for forensic and diagnostic purposes, including the detection of mutations in patient samples and DNA fingerprinting in criminal investigations. It has also been used for gene discovery and mapping.
Southern blotting vs Northern blotting
Although nucleic acid sequences can be identified using either technique, Northern blotting is used to find RNA sequences, and Southern blotting to find DNA sequences. Each involves the same procedures, including gel electrophoresis, membrane transfer, and hybridization.
References
https://www.sciencedirect.com/topics/neuroscience/southern-blot